Project
Fungal infections remain a leading cause of morbidity and mortality for hospitalized patients, since both accurate clinical diagnosis and subsequent therapy pose challenging problems. The central idea of InnateFun is to employ mechanisms of innate immune defense for therapeutic interventions to better treat fungal infections. In a complementary approach InnateFun will design immune-modulatory compounds (IMC) supporting immune surveillance to improve clearance of invasive infections.
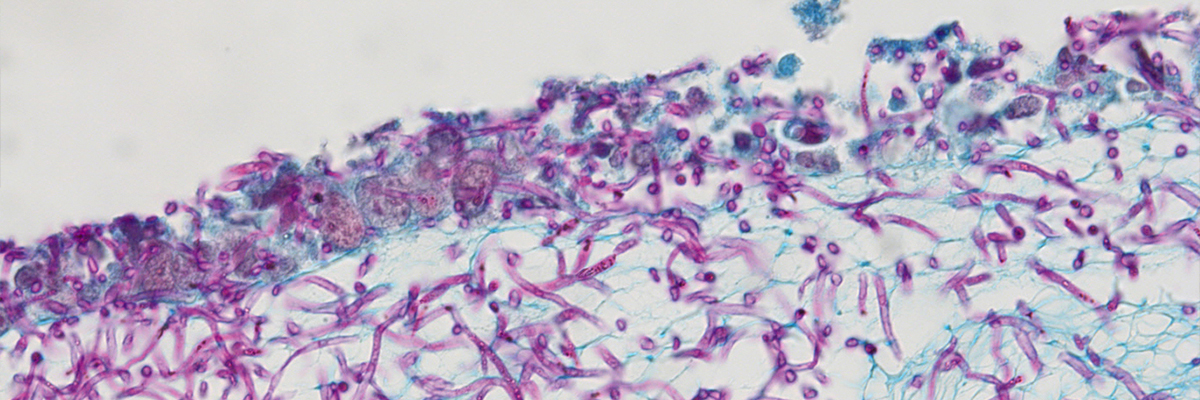
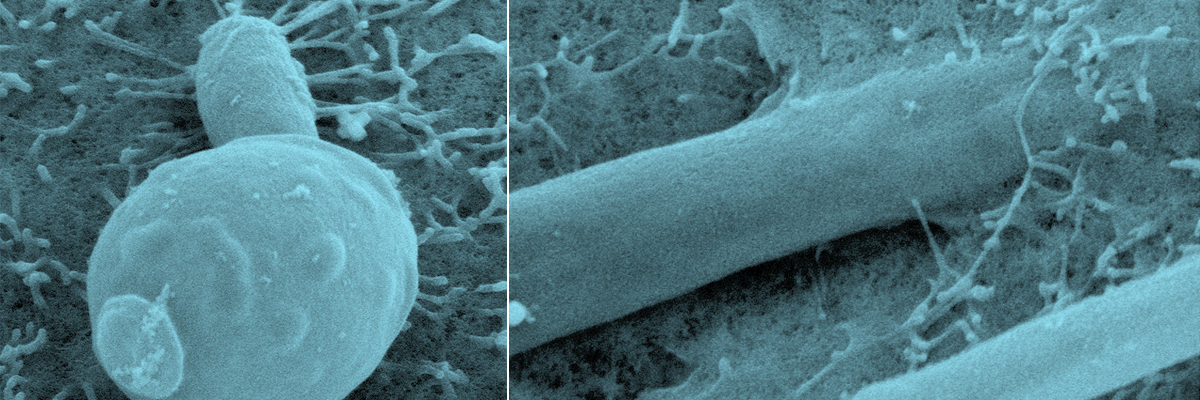
The identification of the novel IMCs will be possible by combining computational methods and iterative cycles of experimental validation to select for IMC-potency on the basis of their ability to modulate cognate host-receptor responses. Stimulating the host-derived immune defense should also be effective against drug-resistant pathogens and against biofilms frequently seen in clinical settings. A major part of invasive fungal infections is caused by Candida spp which are present as normal commensal colonizers on epithelial barriers. Moreover, Candida spp forming biofilms on indwelling medical devices promote fungal persistence and resistance to antifungal agents.
Currently, existing antifungal treatments target growth of fungal pathogens. In contrast, InnateFun aims to modulate innate immune surveillance mechanisms using new IMCs to combat fungal infections. For autoimmune disorders, first medications are already on the market, which target defined components of the immune system. For example, therapeutics effective against Psoriasis have been developed to neutralize cytokines via monoclonal antibodies (acting against IL-17 (Cosentyx) and IL-12 (Stelara)). InnateFun expects to establish a proof-of-principle for a novel type of immune-modulatory antifungal drugs.
Our key aims are to:
i) identify lead IMCs modulating immune defense by targeting receptors governing immune surveillance and fungal clearance
ii) validate selected IMCs using human-based in vitro models and animal models
iii) identify mechanisms critical for innate host immune defense, focusing on epithelial barriers and cellular immunity
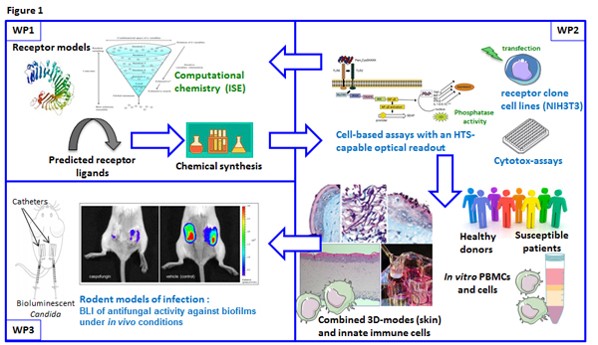
To achieve these aims a work program consisting of three work-packages (WPs) was set up. These work-packages focus on computational chemistry and chemical synthesis (WP1), in vitro/ex vivo screening and validation models (WP2), and validation in rodent models of infection (WP3).
Work-packages
1. Computational Chemistry and Synthesis
- In silico screening (date: month 1 – 12; duration: 12 month)
- Synthesis of IMCs (date: month 6 – 36; duration: 31 month)
- Optimization (date: month 12 – 30; duration: 19 month)
2. Intro/ex vivo Screening Systems
- Setting up assays (date: month 1 – 9; duration: 9 month)
- Identification od IMCs (date: month 1 – 21; duration: 21 month)
- Validation 3D-models (date: month 1 – 30; duration: 30 month)
3. Compound evaluation in rodent models
- Epithelial/Intestinal (date: month 12 – 36; duration: 25 month)
- Biofilm models (date: month 1 – 36; duration: 36 month)
- Systemic models (date: month 12 – 36; duration: 25 month)
Milestones
In addition to the work-packages we have defined six milestones on the way to the achievement of our goals:
M 1.1: First in silico identification of IMCs (against 2/3 of the targets)
M 1.2: Second generation IMCs available (as compounds)
M 1.3: Novel IMCs synthesized (at gram levels for use in rodent models)
M 2.1: Identification of 6-12 new IMCs (in cell based assays)
M 2.2: 4-3 IMCs selected for in vivo validation (based on 3D-models)
M 3.1: IMCs protecting against infection (at least one)